Elisaveta Miladinova
Email: e_miladinova@mail.bg
© 2019 Sift Desk Journals. All Rights Reserved
VOLUME: 3 ISSUE: 1
Page No: 252-260
Elisaveta Miladinova
Email: e_miladinova@mail.bg
Elisaveta Miladinovaa, b*
a Faculty of Physics, Sofia University "St. Kliment Ohridski", 5 James Bourchier Blvd., 1164 Sofia, Bulgaria
b Section of Biosimulation and Bioinformatics, Center for Medical Statistics, Informatics and Intelligent Systems, Medical University of Vienna, Spitalgasse 23, 1090 Vienna, Austria
James A Platts(platts@cardiff.ac.uk)
Hamid Rashedi(hrashedi@ut.ac.ir)
Thomas Olof Sandberg(tsandber@abo.fi)
Christophe J Moreau(christophe.moreau@ibs.fr)
Elisaveta Miladinova, Molecular dynamic study of the stability of oxytocin - divalent zinc complex in aqueous solution(2019) SDRP Journal of Computational Chemistry & Molecular Modelling 3(1)p:252-260
Oxytocin (OT) is a neurohypophysial hormone, which acts both on the peripheral tissues (hormonal), and as a neurotransmitter in the brain. It plays an important role in the control of the uterine contractions during labor, secretion of milk and in many social and behavioural functions. OT accomplishes its functions via interaction with specific oxytocin receptors (OTRs), which belong to the rhodopsin type (class I) group of G-protein coupled receptors (GPCRs). Data from experimental and theoretical studies have shown that the binding of divalent zinc ion to the oxytocin molecule drastically alters its conformation and ensures its correct location in the active center of the receptor. The formation of a stable octahedral complex between oxytocin and divalent zinc ion in an aqueous solution is an important condition necessary for the binding of the hormone to its receptor. So far however, in crystallographic databases there isn't yet information about the 3D structure of the complex.
The aim of our study was to create a rigid structure between the human hormone oxytocin and divalent zinc ion and confirmation of its stability by molecular dynamics (MD) simulations. To achieve this goal a computer model of the hormone - metal complex was built by bonded approach. We used CHARMM 36 force field and artificial bonds, to produce a structure with octahedral orientation of the six carbonyl backbone oxygens of the oxytocin molecule, surrounding the zinc ion. The Gromacs software package was used to check the stability of the constructed oxytocin - zinc ion system by conducting 50 ns simulations in aqueous solution.
Keywords: bonded approach; divalent metal ions; G-protein coupled receptors (GPCRs); molecular dynamics (MD) simulations; oxytocin (OT); oxytocin receptor (OTR).
List of Abbreviations
|
OT |
Oxytocin |
|
OTR |
Oxytocin Receptor |
|
GPCR |
G - Protein Coupled Receptor |
|
MD |
Molecular Dynamics |
|
TM |
Transmembrane |
|
EL |
Extracellular Loop |
|
IL |
Intracellular Loop |
|
IMS – MS |
Ion Mobility Spectrometry - Mass Spectrometry |
|
RMSD |
Root Mean Square Deviation |
|
RMSF |
Root Mean Square Fluctuation |
|
Rg |
Radius of gyration |
|
SASA |
Solvent Accessible Surface Area |
Oxytocin is a nonapeptide (Cysteine-Tyrosine-Isoleucine-Glutamine-AsparagineCysteine-Proline-Leucine-Glycinamide), synthesized by neurons located in the supraoptic and paraventricular nuclei of the hypothalamus, which is secreted toward the blood circulation through the posterior pituitary part in response to various stimuli. The first amino acid residue Cys1 and sixth amino acid residue Cys6 in the oxytocin molecule are connected with a disulfide bridge, which separates the hormone in two parts - disulfide ring and acyclic tripeptide tail (Figure 1A). The central actions of oxytocin are mediated via oxytocin receptors (OTRs) distributed widely in the brain and peripheral tissues. The OTRs belong to the G protein coupled receptors superfamily.
The GPCRs are constructed from seven hydrophobic transmembrane α- helices (TM1-
7) connected by alternating extracellular (EL1, EL2 and EL3) and intracellular loops (IL1, IL2 and IL3). The N-terminal part of their polypeptide chain is located in the extracellular space and the C-terminal part - in the cytosol (intracellular fluid around the organelles) (Figure 1B). Binding of OT to the transmembrane part of OTR activates the receptor which subsequently activates various intracellular signal pathways, triggering numerous effects of the hormone, including contraction of the miometrial cells in uterine smooth muscle. At the interaction of OT with its receptor, both parts of the hormone, cyclic portion (N-terminus) and acyclic tripeptide tail (C-terminus), are bound to two different extracellular loops of the oxytocin receptor. Between the N-terminus of OT and a conserved acidic residue on the receptor there is formed a salt bridge [1, 2].
To exert its physiological functions, OT needs to be activated or biotransformed by metal ions. The natural state of OT is its protonated form, in which the N-terminus amino group is buried inside the molecule of the peptide. In this state the hormone cannot bind successfully with its receptor to produce its physiological functions. As the resulting structures remain the same monovalent metal ions cannot activate OT. But, divalent metal ions like Zn2+ are able to induce a dramatic conformational change in the oxytocin. They transform the nonapeptide in an extended structure with the amino group exposed to the outer surface of the molecule. This allows effective interaction of the N-terminus of oxytocin with the acidic residue of the receptor [2].
The interaction between OTR and its natural ligand oxytocin mediated by metal ions as cofactor is not understood on the molecular level, because the correct conformation of both structures (ligand and receptor) is unknown. And this conformation is a key factor which controls the ligand binding in the biomolecular systems.
In the classical MD simulations most of the widely used force fields do not have appropriate parameters for metal ions. Therefore, for modelling zinc and other divalent ions, that are embedded in peptides, two different methods are developed and generally used to describe the interactions between metal ions and coordinated residues : nonbonded [3] and bonded [4] . In the non-bonded models the atoms are bonded and angled to the central atom, but there are no bonds to the ligands, which means that the metalligand complex can change its configuration when it is in solution. In contrast of this the “bonded model” of the Zn – ligand interactions employ covalent (artificial) bonds between Zn2+ and its ligands to maintain the selected Zn2+ coordination geometry in peptides during simulations in water solution [5].
Obtaining of a stable complex between oxytocin and divalent zinc ion in an aqueous solution is an important condition necessary for the hormone-receptor binding. By using a combination of experimental procedures, like ion mobility spectrometrymass spectrometry (IMS-MS), and theoretical methods, Liu and coworkers, show that when Zn2+ binds to OT, it induces a significant structural change in the molecule of the hormone and this leads to increased receptor affinity for the OT - Zn2+ complex [6, 7].
But their research did not report data for the stability of this complex in water solution.
The aim of this study is to construct a rigid structure between the human hormone oxytocin and the divalent zinc ion and check obtained new complex for the stability in aqueous solution by using the simulation technique molecular dynamics. The created computer model will be useful on molecular level to investigate mechanism of interaction between oxytocin and its receptor.
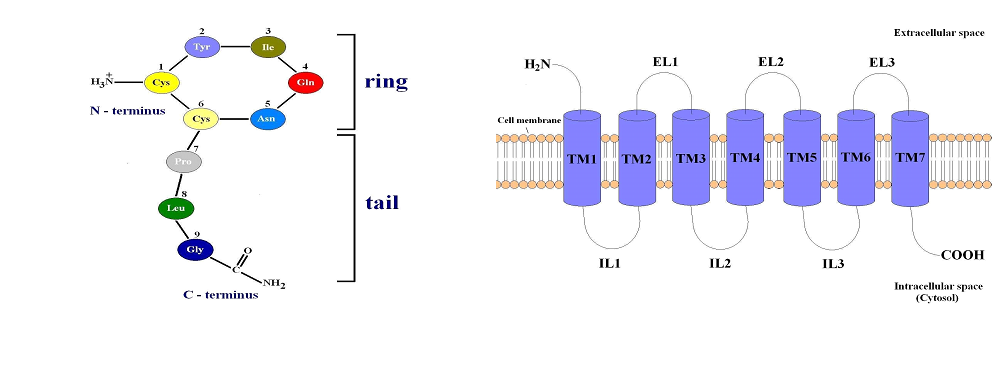
Figure 1. (A) Structure of the human hormone oxytocin. (B) GPCRs structure.
"Bonded approach" used to model zinc and its ligand sphere in oxytocin molecule
In the present study, a model of hormone – metal ion (Zn2+) was created based on the NMR solution structure of oxytocin (Protein Data Bank code 2MGO) [8, 9]. Before the model was constructed the carboxyl function group of Gly9 in OT was substituted with an amide group. We used a bonded model for obtaining the rigid structure between OT and Zn2+ ion. This technique enforces the chosen coordination mode, which prevents exchange or permutation of ligands, or changes in coordination during the simulation.
The artificial bonds [5] between the metal ion and ligands (six of the backbone carbonyl oxygens, associated with Tyr2, Ile3, Gln4, Cys6, Leu8, and Gly9 on the OT) were introduced with bond length restraints and appropriate harmonic potentials.
The restraints of the distance, between backbone carbonyl oxygen atoms of the OT and Zn2+ ion, was used for reducing the system degrees of freedom, letting the complex move as a rigid body.
Molecular dynamics
The stability of the obtained OT - Zn2+ structure in aqueous solution was studied by molecular dynamics. The simulations were performed with GROMACS 5.0.7 package [10], with CHARRMM 36 force field [11]. The OT - Zn2+ system was solvated in cubic box with minimal distance between the peptide and the box walls from 1.2 nm as the solvent was described with the TIP3P water model [12]. The protocol of the simulation procedure comprised initial energy minimization, followed by equilibration and a production run of 50 ns. The minimization was carried out by using the steepest descent algorithm. The duration of the equilibration simulations (NVT and NPT ensembles) was 0.5 ns. All bonds were constrained by LINCS algorithm [13] and a leap-frog integrator was employed with a time-step of 2 fs. Periodic boundary conditions were applied in all directions. The temperature was kept at 310 K with a v-rescale thermostat [14]. The pressure was fixed at 1 bar by means of Parrinello-Rahman barostat [15]. The particlemesh Ewald [16] method was used for calculation of long-range electrostatic interactions. The van der Waals interactions were smoothly switched off at 10-12 Å by a force-switching function [17]. Two separate systems oxytocin with zinc ion and oxytocin without zinc ion was then subjected to molecular dynamics (MD) simulation (50 ns) with the same settings of parameters.
Molecular modelling
A combination of experimental and theoretical evidence indicates that divalent metal ions are the key mediator for the successful binding between oxytocin and its receptor. The oxytocin molecule undergoes substantial conformational change when coordinating with Zn2+, which allow interaction of specific residues in hormone with specific residues in oxytocin receptor [2, 6]. As transition metal the divalent zinc cation has a d10 closed shell. This electron configuration allows different coordination patterns (four-, five- or six-ligand zinc coordination complexes) in water solution. The stable configuration for the zinc divalent cation is the tetrahedral coordination with only four ligands because, its electronic structure favorably accommodates four pairs of electrons in its vacant 4s4p3 orbitals. Experimental studies of five- and six-ligand complexes show that, from energy view point, they are unstable in water solution [18, 19] . Therefore we used the bonded approach to obtain rigid structure between the six carbonyl oxygen atoms from the oxytocin molecule (ligands) and divalent zinc ion. Through this method, the coordinate center is almost frozen, which allows the coordination number to be well-maintained during the whole molecular dynamics simulation. The metal - oxygen bond length was fixed at 2.12 Å. The constructed model is shown in Figure 2 A, B.
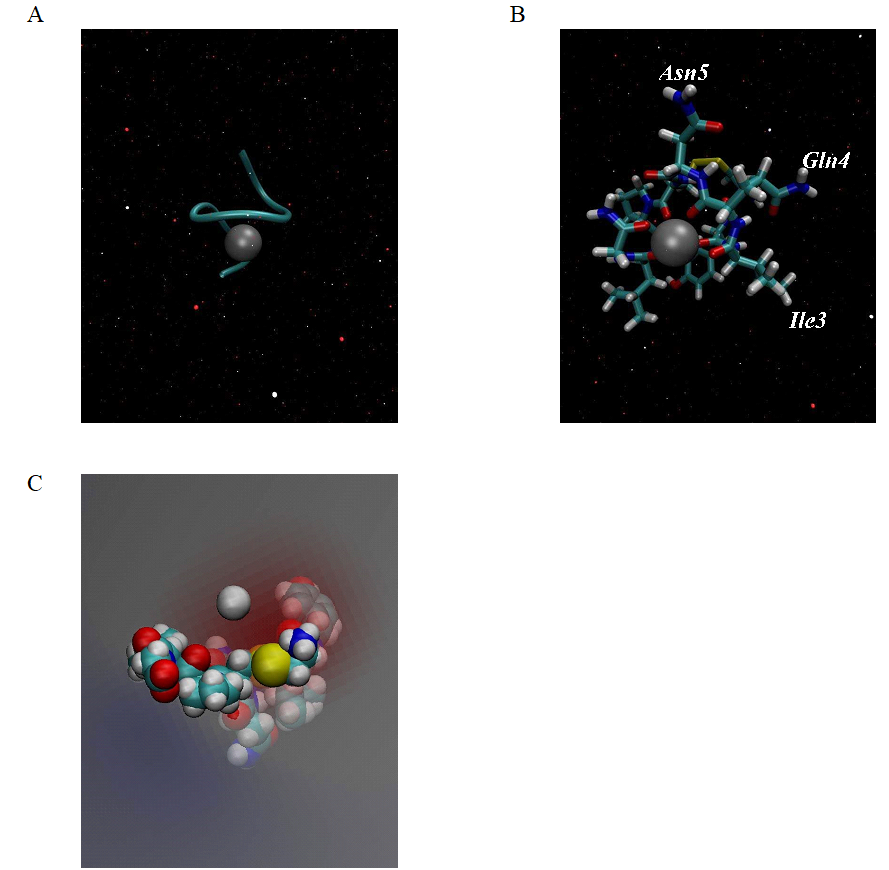
Figure 2. (A) Model of oxytocin - zinc complex, obtained using artificial bonds and Charmm 36 force field. (B) Octahedral orientation of the six carbonyl backbone oxygens of the oxytocin molecule surrounding the Zn2+ ion. (C) Zinc ion in the electric field created by the six oxygen atoms in the oxytocin molecule.
Our model of the 3D structure of oxytocin - divalent zinc ion show that this complex adopts a helical shape (Figure 2A), in which the backbone oxygen atoms of six residues (Tyr2, Ile3, Gln4, Cys6, Leu8 and Gly9) solvated the Zn2+ ion in an octahedral coordination, which is in agreement with previous experimental and computational studies [6]. This means that the Zn2+ ion is tightly embedded in the structure of
oxytocin. The correct folding, like α - helix in our case, of the polypeptide chain is a main condition for many peptides to be activated to accomplish their physiological functions. In biological systems the natural state of oxytocin is its protonated form [OT + H]+ with the amino group of N-terminus buried inside the molecule of the hormone and is physiologically inactive. In the presence of Zn2+ in aqueous solution the natural state of OT is changed to the [OT + M]2+ form, which is with extended structure and, in which the amino group is exposed at the outer surface of the molecule allowing its effective interaction with acidic residue of the oxytocin receptor [2, 6]. In our model the amino group of the N-terminus is in the same position, typically for the [OT + M]2+ form. We also observed that, the coordination of Zn2+ with OT change the spatial orientation of the side chains of Ile3, Gln4, and Asn5 in peptide molecule to form a cohesive, near-planar surface suitable for coordination with the receptor, which is in excellent agreement with the computational findings in [6] (Figure 2B). Since the carbonyl oxygens are orientated toward the interior of the oxytocin molecule they are able to interact with the Zn2+, the surface of [OT + Zn]2+ complex becomes hydrophobic, which is the main condition for desolvation of oxytocin and contributes to binding toward the receptor. The hydrophilic part of the hormone is its tripeptide tail (Pro7-Leu8-Gly9). This shows that the oxytocin is a molecule with an amphiphilic character. Since the six carbonyl oxygen atoms are charged, they generate a negative electrostatic field for the initial binding of Zn2+ to the oxytocin molecule, as is shown in Figure 2C.
Molecular dynamics simulations
The conformational stability, of obtained model of OT - Zn2+ complex, was confirmed by conducting of 50 ns molecular dynamics simulation in aqueous solution. For this purpose, we prepared two systems of study in solution - oxytocin without and oxytocin with zinc ion, to compare the conformational dynamics of peptide hormone in both cases. The Gromacs tools were used for the analyses of the trajectories obtained from the simulations. The following parameters were calculated as a root mean square deviation (RMSD), root mean square fluctuations (RMSF), radius of gyration (Rg) and solvent accessible surface area (SASA). The obtained results were visualized using Xmgrace [20] 2D plotting tool and VMD [21] software.
The root mean square deviation (RMSD) of the backbone of the peptide hormone (without and with zinc ion) is 3.40 Å and is shown in Figure 3A. This big difference in value in the parameter is due to formation of α-helical conformation after binding of the zinc ion toward the oxytocin molecule. This allows its activation by transformation of the ring part of peptide hormone in hydrophobic core suitable for interaction with the receptor. The deviation at the position in backbone atoms of the oxytocin (without and with zinc ion) can be seen on Figure 3B. The graph clearly shows that the native peptide without metal ion has a substantially larger RMSD value (more flexible structure) compared with this of the native peptide with zinc ion in its structure. This means that the zinc ion binding induces significant structural changes in the peptide backbone, which results in its stabilization in aqueous solution. For study of the rigidity of the obtained oxytocin - zinc complex we also made evaluation of the root mean square fluctuations (RMSFs) of the residues of the oxytocin molecule in both cases - without and with zinc ion. The results are presented in Figure 3C. The zinc coordinated peptide is more stable with least fluctuations in comparison to the structure of the peptide without zinc ion, which is characterized with more fluctuation in residues. These observations indicate stabilization of conformational structure of the hormone under influence of the zinc divalent ion. Our results also confirm the weaker mobility of the amino acid residues Tyr2, Ile3, Gln4, Cys6, Leu8 and Gly9 of the oxytocin - zinc complex compared to the structure of the hormone without metal ion. From these six residues the most flexible is Gly9 for the both compared structures of oxytocin (Figure 3C).
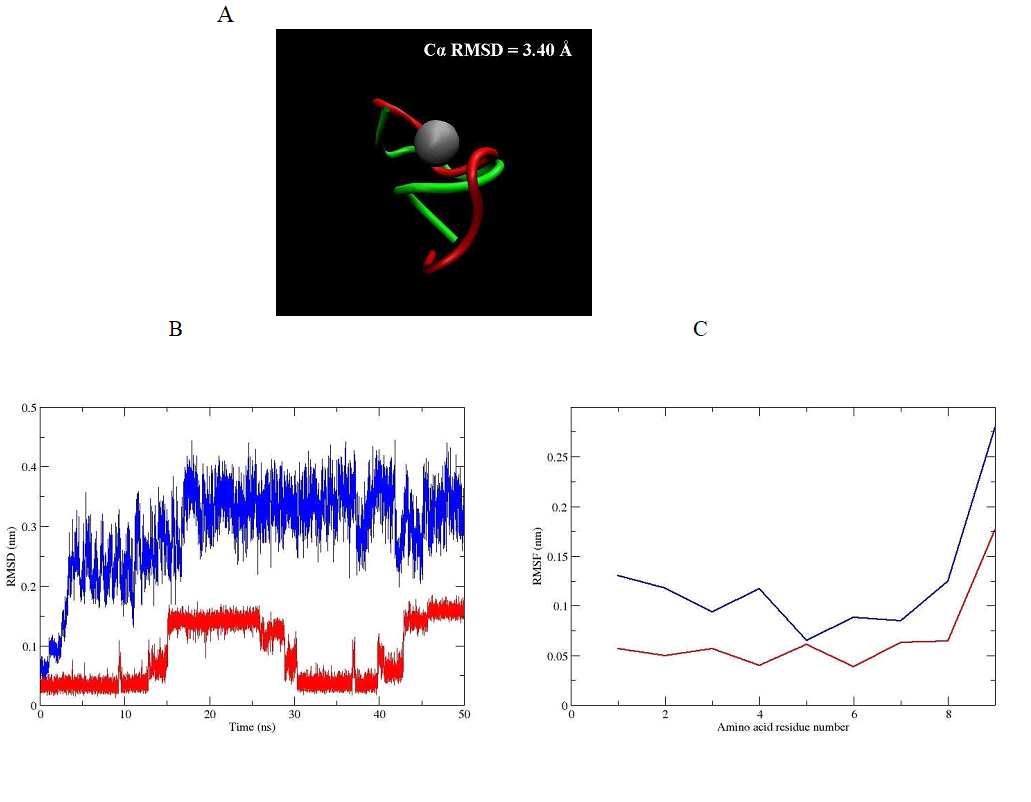
Figure 3. (A) Structural superimposition of the conformation of obtained oxytocin - zinc complex by Cα RMSD value (in green) and the structure of oxytocin - without zinc ion (in red). (B) RMSD of the backbone atoms and (C) RMSF of the amino acid residues for oxytocin without zinc ion (blue line) and oxytocin - zinc complex (red line) during the classical MD simulations.
The stabilization of the more compact peptide conformation, which is exactly that associated with a divalent zinc ion, can be seen of the radius of gyration (Rg), shown in Figure 4A. The Rg plot for the peptide without zinc ion demonstrates a more open conformation. The same trend was observed when total solvent accessible surface area (SASA) was evaluated. The SASA parameter is commonly used for peptides as a measure of the free energy of the solvating polar and non-polar groups. Our results (Figure 4B) revealed, that this area accessible for the water molecules decreased for oxytocin - zinc complex, and increased for the molecule of the hormone with missing zinc ion. As can be seen from the graph represented in Figure 4C, SASA is with lower value for the amino acid residues in position 1, 2, 4, 6, 7 , 8 and 9 (exception for Ile3 - higher value) for the oxytocin - zinc complex and with higher value for the residues in positions 1, 2, 4, 7, 8 and 9 (exception for Ile3 - low value) for zinc free oxytocin molecule. The evaluated parameter is with the same value only for Cys6 and in both cases. The obtained results confirm the conformational stability of our model of oxytocin - zinc comlex in water solution.
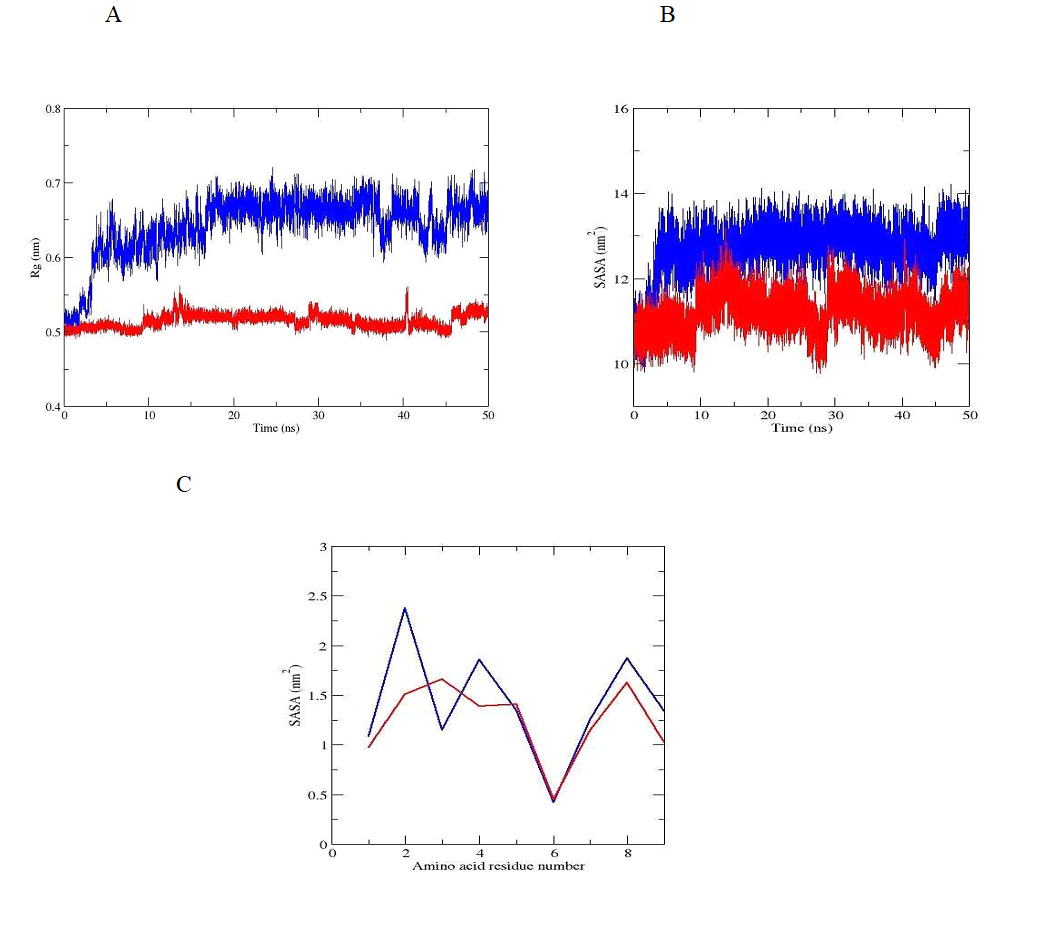
Figure 4. Radius of gyration, (B) SASA and (C) SASA of the amino acid residues of oxytocin without zinc ion (blue line) and oxytocin - zinc complex (red line) during the classical MD simulations.
In this study are shown for first time results, obtained by molecular dynamics simulations, that confirm the stability of the octahedral structure of oxytocin - divalent zinc complex in aqueous solution. An important condition for the successful binding between oxytocin and its receptor is the transformation of oxytocin from biologically inactive into biologically active form, with a stable six-fold coordination of the zinc ion in the native peptide molecule. Since, at physiological pH in aqueous solution, the sixcoordinated peptide complexes are energetically disadvantageous and therefore unstable, we used a bonded approach for creating a new rigid complex (3D model), between oxytocin and a zinc ion. In our computer model the binding of divalent ion by six covalent (artificial) bonds to the oxytocin molecule drastically alters its conformation (in octahedral), as thus ensures its correct location in the active receptor site. This substantial change is visible clearly from the high value of the parameter RMSD. The RMSD of the Cα atoms between the oxytocin - zinc complex and the structure of oxytocin – without zinc ion is 3.40 Å. The helical shape, which adopts our model is with spatial orientation of the side chains of Ile3, Gln4, and Asn5 in peptide molecule, which is suitable for coordination with the oxytocin receptor, which is in agreement with other theoretical findings, as well as experimental data [6]. Our results show, that the obtained structure of oxytocin - zinc complex stay with a stable six-fold coordination of the metal ion in the molecule of the peptide hormone, during the 50 ns simulation. This in turn means a stability of conformation of the oxytocin-zinc complex in aqueous solution as opposed to the high mobility of the oxytocin molecule not associated with zinc ion.
Our model of the oxytocin - zinc complex is suitable for the study of the mechanism of the binding between oxytocin and its receptor and can be used as a template for creating new highly selective antagonists to prevent premature birth.
MD simulations were performed at the parallel cluster of the Atomic Physics Department at Faculty of Physics of Sofia University “St. Kl. Ohridski”. This work was supported in part by the CEEPUS programme and Austrian Agency for International Cooperation in Education and Research (OeAD - GmbH). Thanks to funding by this programme, some of the studies were conducted at the Center for Medical Statistics, Informatics and Intelligent Systems, Medical University of Vienna, Austria. The author thanks Prof. Wolfgang Schreiner and his group and also Assoc. Prof. Thomas Stockner for their help. I would like to thank Georgi Miladinov for creating Fig. 1.
Vrachnis, N., Malamas, F., Sifakis, S., Deligeoroglou, E., Iliodromiti, Z. The Oxytocin - Oxytocin Receptor System and Its Antagonists as Tocolytic Agents. Int J Endocrinol 2011 (2011) 1-8.
View ArticleXu, X., Yu, W., Huang, Z., Lin, Z. Comprehensive Density Functional Theory Study on the Mechanism of Activation of the Nonapeptide Hormone Oxytocin by Metal Ions. J. Phys. Chem. B 114 (2010) 1417-1423. doi: 10.1021/jp907436p.
View ArticleStote, R. H., Karplus, M. Zinc binding in proteins and solution: A simple but accurate nonbonded representation. Proteins: Structure, Function, and Genetics 23 (1995) 12-31.
View ArticleHoops, S. C., Anderson, K. W., Merz, K. M. Force field design for metalloproteins. J. Am. Chem. Soc. 113 (1991) 8262-8270.
View ArticleBrandt, E. G., Hellgren, M., Brinck, T., Bergman, T., Edholm, O. Molecular dynamics study of zinc binding to cysteines in a peptide mimic of the alcohol dehydrogenase structural zinc site. Phys. Chem. Chem. Phys. 11 (2009) 975-983. doi: 10.1039/b815482a.
View ArticleLiu, D., Seuthe, A., Ehrler, O., Zhang, X., Wyttenbach, T., Hsu, J., Bowers, M. Oxytocin - Receptor Binding: Why Divalent Metals Are Essential. JACS Communications 127 (2005) 2024-2025. doi: 10.1021/ja046042v.
View ArticleWyttenbach, T., Liu, D., Bowers, M. T. Interactions of the hormone oxytocin with divalent metal ions. J. Am. Chem. Soc. 130 (2008) 5993-6000.
View ArticleBernstein, F., Koetzle, T., Williams, G., Meyer, E., Brice, M., Rodgers, J., Kennard, O., Shimanouchi, T., Tasumi, M. The Protein Data Bank: a computer-based archival file for macromolecular structures. J. Mol. Biol. 112 (1977) 535-542. 80200-3
View ArticleKoehbach, J., O'Brien, M., Muttenthaler, M., Miazzo, M., Akcan, M., Elliott, A., Daly, N., Harvey , P., Arrowsmith, S., Gunasekera, S., Smith, T., Wray, S., Goransson, U., Dawson, P., Craik, D., Freissmuth, M., Gruber, C. Oxytocic plant cyclotides as templates for peptide G protein-coupled receptor ligand design. Proc. Natl. Acad. Sci. U. S. A. 110 (2013) 21183-21188.
View ArticleAbracham, M. J., Murtola, T., Schulz, R., ..., Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelizm from laptops to supercomputers. Software X 1-2 (2015) 19-25.
View ArticleHuang, J., MacKerell, A. D. CHARMM36 all-atom additive protein force field: validation based on comparison to NMR data. J Comput Chem 34 (2013) 2135-2145.
View ArticleJorgensen, W., Chandrasekhar, J., Madura, J., Impey, R., Klein, M. Comparison of simple potential functions for simulating liquid water. J CHEM PHYS 79 (1983) 926935.
View ArticleHess, B., Bekker, H., Berendsen, H., Fraaije, J. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 18 (1997) 1463-1472. 1096-987X(199709)18:12<1463::AID-JCC4>3.3.CO;2-L
View ArticleBussi, G., Donadio, D., Parrinello, M. Canonical sampling through velocity rescaling. J CHEM PHYS 126 (2007). 17, 1302 (1978).
View ArticleParrinello, M., Rahman, A. Polymorphic transitions in single crystals: A new molecular dynamics method. J APPL PHYS 52 (1981) 7182-7190.
View ArticleEssmann, U., Perera, L., Berkowitz, M. L., Darden, T., Lee, H., Pedersen, L. G. A smooth particle mesh Ewald method. J CHEM PHYS 103 (1995) 8577-8593.
View ArticleSteinbach, P. J., Brooks, B. R. New spherical-cutoff methods for long-range forces in macromolecular simulation. J COMPUT CHEM 15 (1994) 667-683.
View ArticlePang, Y.-P., Xu, K., Yazal, J., Prendergast, F. Successful molecular dynamics simulation of the zinc-bound farnesyltransferase using the cationic dummy atom approach. Protein Science 9 (2000) 1857-1865. PMID: 11106157.
Roe, R. R., Pang, Y. -P. Zinc′s Exclusive Tetrahedral Coordination Governed by Its Electronic Structure. J MOL MODEL 5 (1999) 134-140.
View ArticleTurner, P. J. XMGRACE, Version 5.1.19. Center for Coastal and Land-Margin Research, Oregon Graduate Institute of Science and Technology, Beaverton, OR, (2005).
Humphrey, W., Dalke, A., Schulten, K. VMD: visual molecular dynamics. J Mol Graph 14 (1996) 33-8, 27-8. 00018-5
View Article